
Objective: To study the growth & propionic acid production kinetics of Propionibacterium acidipropionici under batch cultivation conditions in bioreactor.
Theory: Basic concepts & importance of Propionic acid production from renewable resources
Propionic acid fermentation –
Propionic acid has been a product of industrial importance. It is widely used in the manufacture of herbicides, cellulose plastics, perfumes, artificial fruit flavours, mold inhibitors, food and feed preservatives. The Na+, Ca+ and K+ salts of propionic acid have been listed as food preservatives categorized as Generally Recognized As Safe (GRAS) food additives (Colomban et al., 1993).
Presently the commercial production of propionic acid is entirely by petrochemical routes (Jin and Yang 1998). Since the petrochemical reserves are gradually depleting, research efforts are being directed towards the production of propionic acid by alternative renewable resources mainly by fermentation.
Batch propionic acid fermentation takes about 3 days to reach approximately 2 % (w/v) propionic acid. Propionic acid fermentation by P. acidipropionici is a complex multiproduct fermentation. It is rather difficult to optimize such a process with the conventional trial and error approach With the recent advances in bioprocess modeling, together with the knowledge of biochemistry of fermentation, it is possible to formulate an empirical mathematical model which could facilitate better understanding of actual process behavior and ease optimization efforts.
According to the biochemistry of the fermentation the organism P. acidipropionici grows (under strict anaerobic conditions) using lactose as the carbon sources at pH 6.5 and temperature 30ºC.
Modelling of Propionic acid fermentation –
Following were the assumptions for the development of the model –
- Lactose is the only limiting substrate in the batch cultivation
- There is no process limitation by the nitrogen source
- The culture inhibition is by the accumulating metabolic products (Propionic and acetic acids only).
- The pH is known and controlled at a constant value throughout the modeled period.
- Propionic acid production is by direct conversion of lactose as its intermediate succinic acid does not demonstrate accumulation and consumption like pyruvic acid.
The specific substrate consumption rate (rs) featured limitation of substrate (as per the Monod’s type of representation with rsmax as maximum specific substrate consumption rate) lactose and inhibition due to propionic (P1) and acetic acids (P3) (which exhibited exponential decrease of rs by increasing the values of P1 & P3.
The present description of µ overcomes the limitation of traditional Monod’s model which when used with sp substrate consumption rate create serious anomaly where by the culture features consumption of substrate even when the no substrate is actually present in the reactor. The proposed model in fact of returns decreased growth in the absence of zero substrate concentration which is normally observed in the bioreactor.
The propionic acid specific growth rate was assumed to be directly dependent on specific substrate consumption rate in a growth related as well as non growth associated consumption of substrate.
The net pyruvic acid specific growth rate was expressed as difference of production resulting from consumption of lactose and its consumption to produce Acetic & Succinic acid.
The net acetic acid (P3) specific growth rate was assumed to be directly dependent on consumption of substrate together with the contribution of conversion of pyruvic acid.
The net Succinic acid (P4) specific growth rate was assumed to be directly dependent on consumption of substrate together with the contribution of conversion of pyruvic acid.
For estimation of the optimal model parameters, a non-linear regression technique (Bard, 1974) assisted by a computer program (Votruba, 1982; Volesky and Votruba, 1992) was used to minimize the deviation between the model predictions and batch experimental data. For calculation of the model predictions, the system of differential equations for substrate, biomass, propionic acid, pyruvic acid, acetic acid and succinic acid which describe the batch growth and Propionic acid production kinetics, was solved by an integration program based on Runge-Kutta Method of 4th order (Volesky and Votruba, 1992). The optimization program for the direct search of the minimum of a multivariable function was based on the original method of Rosenbrock (Rosenbrock, 1960). The minimization criteria used in the program was as follows:
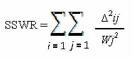
where SSWR represents the “sum of squares of weighed residues”, subscript “i and j” represent the number of experimental data points and the number of variables respectively, Wj represents the weight of each variable (usually the maximum value of each variable) and Δij denotes the difference between the model and experimental values.
The values of the optimized parameters are summarized in Simulator.




